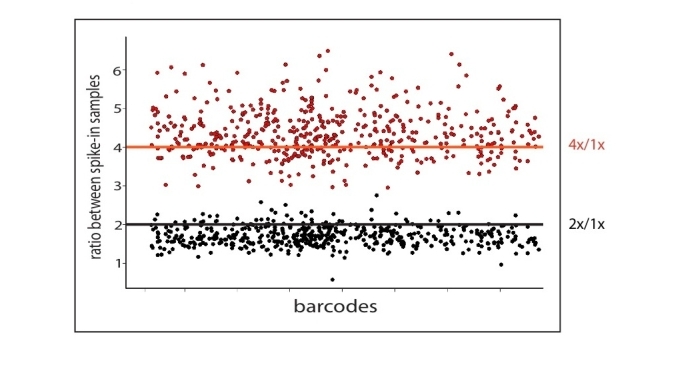
Fig S3 : Signal ratios in Pool-Constant Spike-ins
Strain counts were assessed in a spike-in pool of 953 strains after amplication from three increasing template quantities designated as 4x, 2x and 1x. Strain count ratios (4X/1X in red, 2X/1X in black) were calculated. The horizontal line indicates the expected ratio based on the amount of PCR product added to the sequence reaction. Ratios are ordered across the x-axis by barcode. Counts not exceeding a threshold of 100 in 1x sample were discarded.