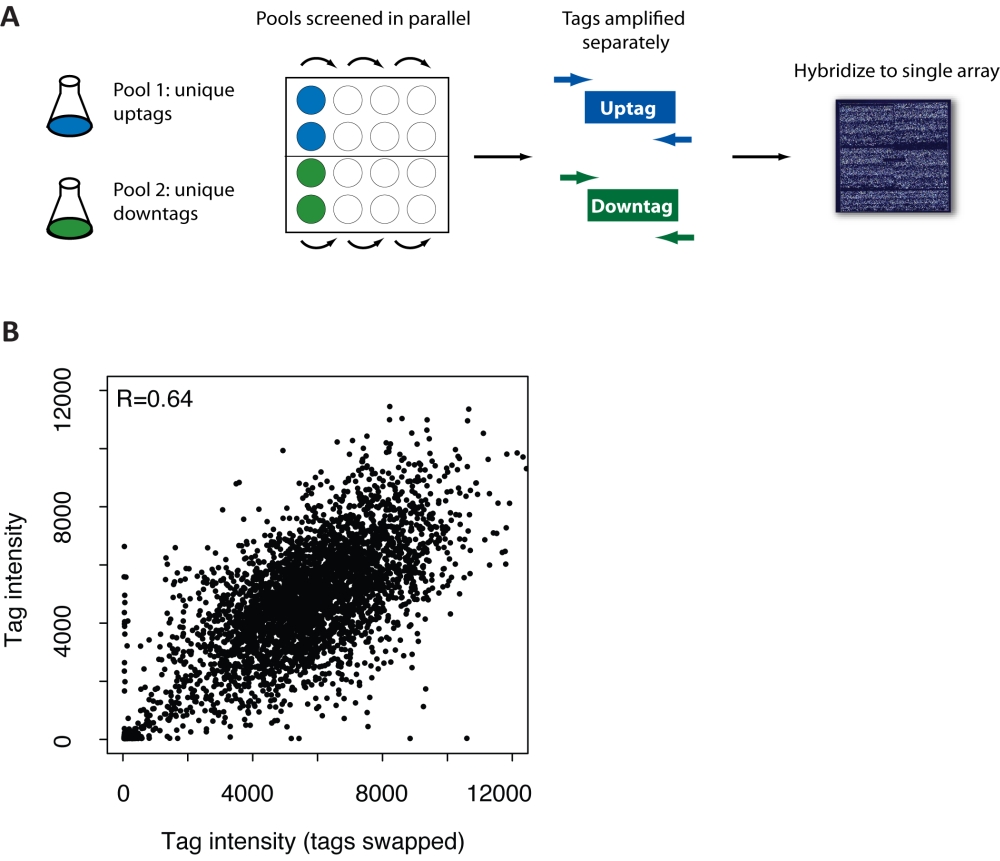
Figure S2: Scheme and validation for using one tag per strain.(A) Unique uptags were selected for one pool and unique downtags for a second pool, overall representing 4401 strains (4388 unique genes). The two pools are then screened and tags amplified in parallel to prevent cross-contamination of overlapping tags. The uptag and downtag PCRs can then be combined prior to hybridization. (B) Validation of the one tag per strain approach outlined in Figure S2A. Two uniquely tagged pools were used to increase the number of strains able to be represented on an array. Hybridization performance of the pool was compared to see if strain tracking was affected depending on whether the uptag or a downtag was used to represent a strain. Two independent pools (“pool 1” and “pool 2”) of the 4252 successfully transformed strains were grown for 20 generations in YPD + 1% DMSO. Uptags were amplified from pool 1, and downtags from pool 2, and hybridized to a TAG4 array. In a “tag swap”, downtags from pool 1 and uptags from pool 2 were then amplified and hybridized to an array. Pearson correlation of tag intensities above 3X background is indicated in the upper left corner. |
 |