Overview
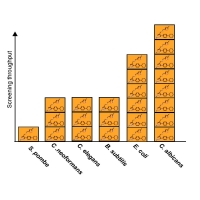
Pre-selection of compounds that are more likely to induce a detectable phenotype can increase the efficiency and reduce the costs for model organism screening. To identify such molecules, we screened ~81,000 compounds in S. cerevisiae and identified ~7,500 that inhibit cell growth. Screening these growth-inhibitory molecules across a diverse panel of model organisms resulted in an increased phenotypic hit rate. This data was used to build a model to predict compounds that inhibit yeast growth. Empirical and in silico application of the model enriched the discovery of bioactive compounds in diverse model organisms. To demonstrate the potential of these molecules as lead chemical probes we performed genome-wide phenotypic screens and identified specific inhibitors of lanosterol synthase and of stearoyl-CoA 9-desaturase. This community resource of growth inhibitory molecules have been made commercially available and the computational model and filter are provided.
Significance
We have presented three approaches, based on yeast growth inhibition, to guide compound selection to reduce the costs associated with model organism screening programs. Screening compounds that are known to inhibit yeast growth enriches for compounds that induce a variety of phenotypes in diverse model organisms, and these compounds, with further optimization, may yield specific chemical probes. Designing a library of compounds based on those that pass a filter based on two physicochemical properties can decrease costs by ~25%, and is easy to implement. Purchasing compounds rank-ordered by their likelihood to result in a desired phenotype based on a Naïve Bayes model can also dramatically reduce costs. One can also take advantage of newly generated screening data to rebuild the model in the context of the model organism of interest. This iterative approach is key because no model will perform optimally in all applications.
As an experimental resource the yactive compounds are available as a pre-plated collection from ChemBridge, and a list of 14M purchasable compounds scored by our Naïve Bayes model and 2-property filter is available for download from this website in the Datasets section (Datasets S12 and S13).