
![]()
![]()
![]()
![]()
![]()
Protocols &Technical Information
![]()
![]()
Confirmation PCR primer Design
The confirmation primers were designed using a modified version of the WI institute PRIMER program, version 2.3. For information and references about this program see the Whitehead Institute Genome Homepage. The primers were designed to have lengths between 17 and 28 bases and to have melting temperatures of 65°C plus or minus 2°C. The minimum GC was set to be 30 % and maximum to be 70%. Primers A and D were picked from regions 200 to 400 bases upstream or downstream, respectively, of the open reading frame. Primers B and C were chosen from regions within the open reading frame and were designed to give PCR products with sizes of 300-1000 bases when used with A or D, respectively.
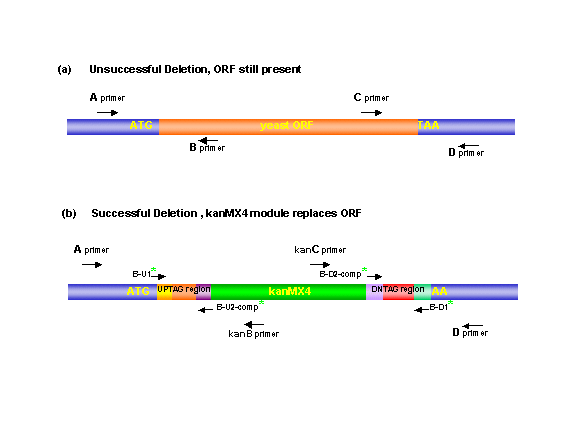
When the primers were chosen, a primer record was created. This file contains
the parameters that were used to select the primers, and A and B , or
C and D primer sequence for each ORF. It also contains the expected PCR
product sizes that would be observed for an A-B (- deletion) or A-KanB
(+deletion) amplification event.
We learned that Primer 2.3, which had been used to pick confirmation primers
for chromosomes V, XIII and I, has a bug in it. The program picked primers
that have melting temps that are lower than the requested melting temperature
(65°C). In reality, the primers produced had melting temps that are
between 53°C and 58°C. The primers still worked extremely well.
We estimated the confirmation PCR is between 90 and 95% percent successful
when performed on wildtype template. However, we did not determine whether
unsuccessful PCR's occured because of primer synthesis failure, template
quality or primer design failure.
![]() last updated August 2002 amchu@cmgm.stanford.edu
last updated August 2002 amchu@cmgm.stanford.edu